©2024 - Stefano Forte
GENOMICS AND EXPERIMENTAL ONCOLOGY UNIT
"Sono la cultura e la storia a dividere i popoli, non la biologia. La biologia ci unisce, e ci unisce anche a molti altri esseri viventi" E. Boncinelli

RESEARCH INTERESTS
Dr. Stefano Forte’s laboratory focuses on uncovering the mechanisms underlying cancer progression and developing predictive models for oncological diseases. Recently, the team has identified an innovative in vitro model based on tumor-initiating cells, which demonstrates the ability to predict the efficacy of radiotherapy in vivo.
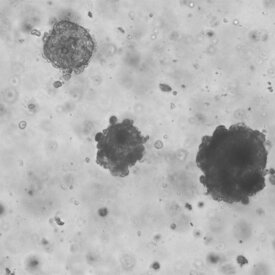
In addition, the lab is actively engaged in developing advanced in vitro oncological models using patient-derived tumor organoids (PDTOs). These models offer a robust platform for personalized medicine by closely mimicking the tumor microenvironment of individual patients. The research also extends to the characterization of signaling pathways mediated by extracellular vesicles, which play a pivotal role in tumor progression and intercellular communication.
A key area of focus for the lab is the application of single-cell RNA sequencing to elucidate the impact of tumor heterogeneity on treatment responses. By analyzing gene expression at the single-cell level, the team aims to unravel the complex interplay between diverse tumor cell populations and their therapeutic vulnerabilities.
By integrating cellular and molecular approaches, the team strives to elucidate the substrates driving these processes, advancing both our understanding of cancer biology and the development of effective therapeutic strategies.
EXPERIMENTAL ONCOLOGY
Every tumor responds to treatment differently. Although numerous studies have been conducted to identify the prerequisites for sensitivity to radiation therapy, the topic remains poorly understood. Locally advanced rectal cancers (LARC) are frequently treated with neoadjuvant radiotherapy or chemoradiotherapy to achieve tumor downstaging. This approach can make subsequent surgery safer and improve remission rates. In some cases, tumors respond exceptionally well, undergoing significant regression. A subset of these tumors may even achieve complete regression, raising the possibility that curative surgery might not be necessary if these cases could be accurately predicted. However, there are instances where tumors fail to respond to neoadjuvant therapy, presenting a significant challenge.
Being able to predict the response to radiotherapy or chemoradiotherapy in advance would enable clinicians to tailor therapeutic strategies to each patient more effectively. This individualized approach could optimize outcomes, sparing patients from unnecessary treatments and focusing resources on approaches more likely to succeed. Furthermore, it is essential to consider the potential toxic effects of radiotherapy on healthy tissues, which are directly proportional to the doses used. Predicting the minimum effective dose for each patient could significantly reduce toxicity while maintaining therapeutic efficacy, thereby improving the overall quality of care.
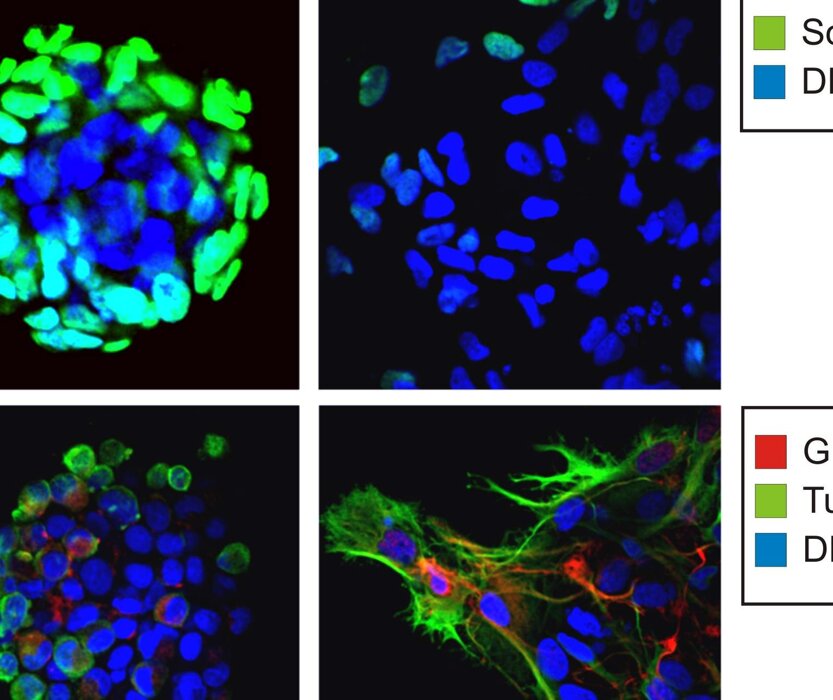
In our laboratory, we have developed an innovative in vitro model capable of predicting the effects of therapy in the cases we have analyzed. Our research has primarily focused on rectal and lung cancers. By isolating cancer stem cells from patient biopsies, we were able to treat these cells in vitro and observe their responses. Notably, we found a correlation between the in vitro response of these cells and the clinical prognosis of the donor patients. For lung cancers, we identified the lowest effective dose needed to eliminate cancer cells in vitro. This dose was consistent with the dose required to achieve a significant reduction in tumor mass in vivo.
In addition to these efforts, we are employing single-cell transcriptomics to further elucidate the impact of tumor heterogeneity on therapeutic responses. This cutting-edge approach allows us to analyze gene expression profiles at the resolution of individual cells, providing deeper insights into the diverse cellular subpopulations within tumors. By understanding how these subpopulations contribute to therapy resistance or sensitivity, we aim to uncover new biomarkers and therapeutic targets. This research highlights the crucial role of tumor heterogeneity in shaping treatment outcomes and underscores the importance of personalized medicine in oncology.
Our findings underscore the potential of personalized approaches in cancer treatment. By leveraging patient-derived cancer stem cells, in vitro modeling, and single-cell transcriptomics, we aim to refine therapeutic strategies, reduce toxicity, and improve outcomes for patients. The ability to predict therapeutic responses and effective dosing represents a significant step forward in the pursuit of precision oncology.

TECHNOLOGIES
©2024 - Stefano Forte


